From mRNA To Protein: Overview of Translation Biology
Get an overview of translation biology – the process where the encoded info in mRNA is translated into amino acids and proteins (Protein Synthesis Steps).
Jump to:
In this article, you will be introduced to the process of protein synthesis, also referred to as translation. Let’s take a very close look.
What is Translation in Biology?
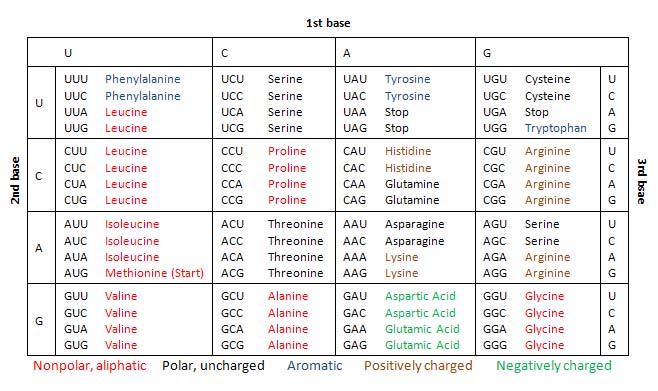
- In particular, the mRNA is read in groups of three bases called codons. Apparently, there is a total of 61 codons that code for 20 specific amino acids.
- The relationship between the mRNA codons and their corresponding amino acids are collectively referred to as the genetic code[2] (shown in the table below).
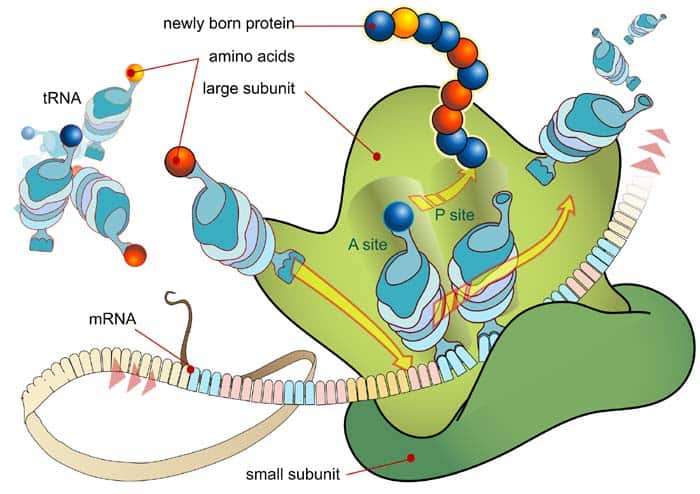
During translation, the mRNA codons are read from their 5′ ends to their 3′ ends by transfer RNA (tRNA), with one of its end having an anticodon that binds with the mRNA base pairing, and anther end carrying the amino acid by the specific codon.
![]()
Where Does Translation Occur?
- In both cases, the ribosomes are composed of two subunits which contain rRNA (ribosomal ribonucleic acid) and proteins.
- In prokaryotes, the small subunit is referred to as the 30S while the large subunit is designated as the 50S. On the other hand, the small and large subunit of the eukaryotic ribosome are designated as 40S and 60S respectively.
![]()
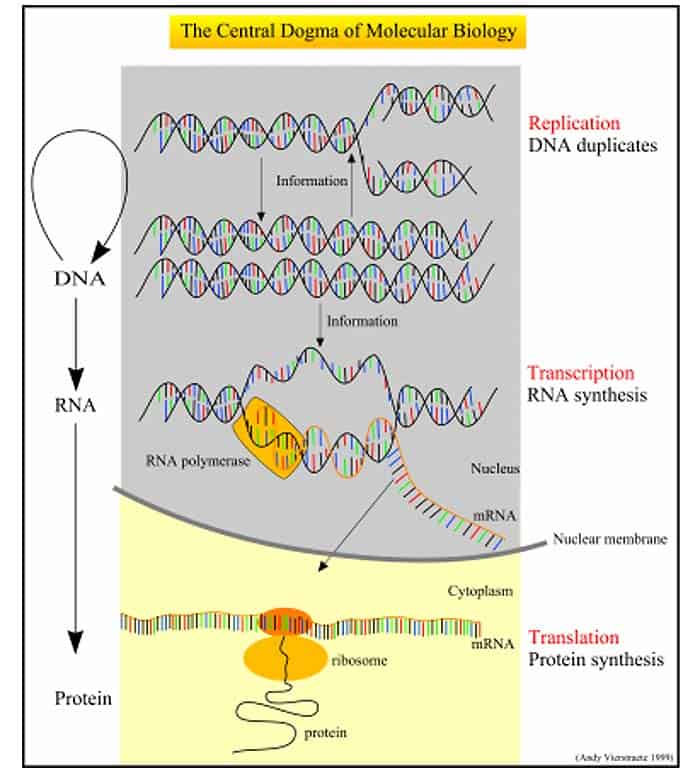
Events Prior To Translation
![]()
The Three Stages of Translation
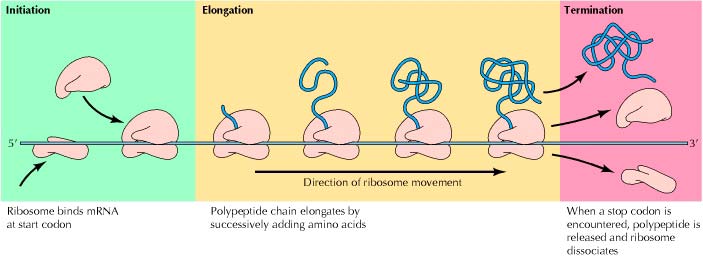
1. initiation
The process of translation is always initiated by the start codon AUG (encodes the amino acid methionine). Alternatively, it can be substituted by the codon GUG (encodes the amino acid valine).
a. Prokaryotic cells
In prokaryotes like bacteria, the initiation of translation begins with the binding of the initiation factors 1, 2, and 3 (IF-1, IF-2, and IF-3) to the 30S ribosomal subunit.
- This is then followed by the binding of the mRNA and initiator tRNA to the complex, triggering the release of the IF-3 to allow the 50S ribosomal subunit to join the previous complex.
- The GTP (guanosine triphosphate) is then hydrolyzed to release IF-1 and IF-2, following the formation of the 70S initiation complex for the next step–elongation.
b. Eukaryotic cells
On the other hand, the process of initiation in eukaryotic cells is more complex and requires more initiation factors. For instance, the eukaryotic initiation factors such as eIF-1, eIF-1A, and eIF-3 attach to the 40S ribosomal subunit, while the eIF-2 joins the initiator methionyl tRNA.
- The eIF-4 factors then search for the mRNA and bring it to the 40S ribosomal subunit.
- Unlike prokaryotic initiation factors, eukaryotic initiation factors can recognize the 5′ and 3′ ends of the mRNA, which is specifically responsible for polyadenylation during translation.
- The 40S ribosomal subunit, together with the associated eIFs and methionyl tRNA the browses the mRNA sequence to look for the start codon (AUG).
- After reaching AUG, eIF-5 triggers the release of both eIF-5 and eIF-2 initiation factors, resulting to the binding of the 40S subunit to the 60S subunit to form the 80S initiation complex unique to eukaryotes.
![]()
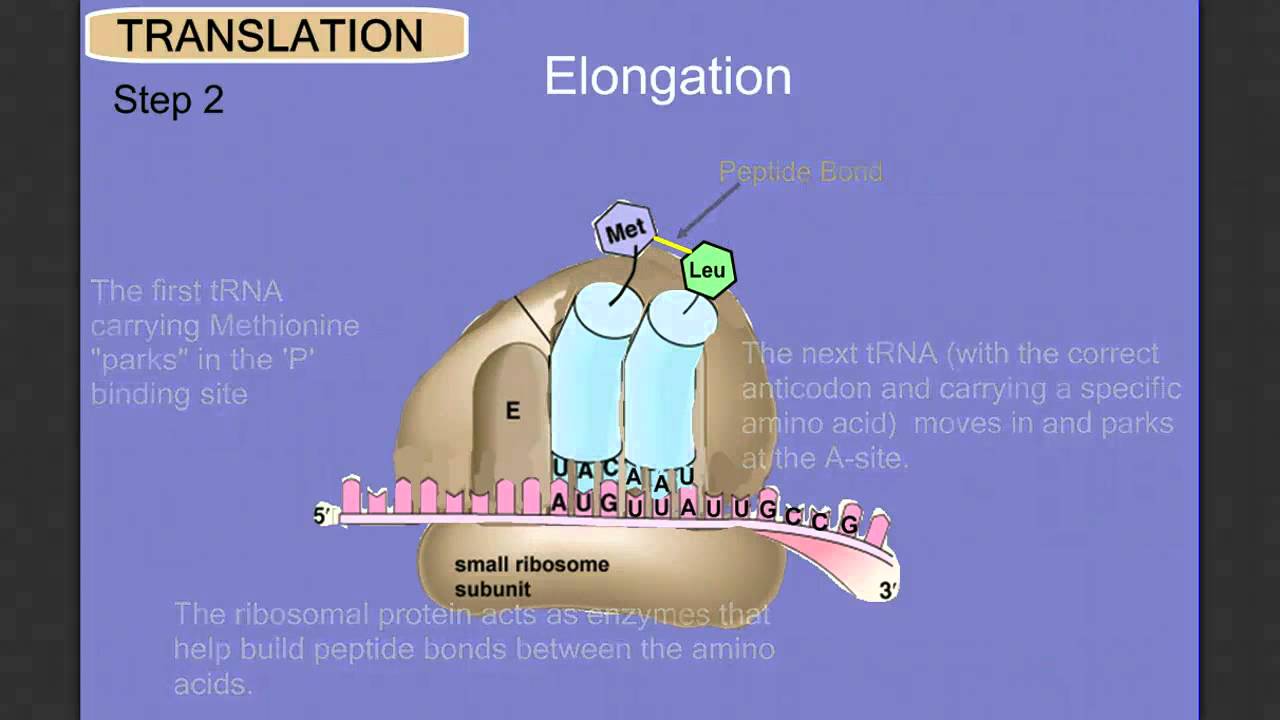
2. Elongation
- The ribosome has three specific sites for the binding of tRNA: aminoacyl (A site), peptidyl (P site), and exit (E site). In particular, the peptidyl (P) site is where the initiator methionyl tRNA binds in order to initialize elongation.
- To formally start elongation, the aminoacyl tRNA binds to the aminoacyl (A) site through pairing up with the second codon of the mRNA sequence.
- After that, elongation factors (EF-Tu in prokaryotic cells; eEF-1α ineukaryotic cells) help the aminoacyl tRNA to the ribosome. GTP must be first hydrolyzed in order to release the elongation factors bound to it.
- It can be observed that spending high amounts of GTP in this particular stage can help make protein synthesis more accurate as it allows more time for proofreading before the peptide bond forms.
- As the process continues, the elongation factors bound to GDP should return to being GTP by using elongation factors EF-Ts.
![]()
3. Termination
- In order to recognize signals and stop the synthesis of proteins, the release factors of cells bind to a stop codon (any of the three mentioned above) at the aminoacyl (A) site and hydrolyze the bond between the chain and the tRNA at the peptidyl (P) site.
- This further results to the releasing of the polypeptide chain from the ribosome, tRNA, and the ribosomal subunits, completely dissociating the mRNA template.
![]()
Indeed, the process of translation is very essential for organisms and we have seen how it works in cells. Now, after knowing how crucial the information is in each stage, what do you think will happen if there are any failures or errors during translation?
![]()
Cite this page
Bio Explorer. (2026, January 6). From mRNA To Protein: Overview of Translation Biology. https://www.bioexplorer.net/translation-biology-protein-synthesis.html/
References
- [1] – “The Central Dogma of Biology: Definition & Theory – Video & Lesson Transcript | Study.com”. Accessed March 27, 2017. Link.
- [2] – “Stages of translation (article) |
Khan Academy”. Accessed March 27, 2017. Link. - [3] – “ribosomes – The Cell – NCBI Bookshelf”. Accessed March 27, 2017. Link.
- [4] – “From DNA to RNA – Molecular Biology of the Cell – NCBI Bookshelf”. Accessed March 27, 2017. Link.
- [5] – “Translation of mRNA – The Cell – NCBI Bookshelf”. Accessed March 27, 2017. Link.