These topics focus on the discoveries and developments in microbiology in 2021. Research and developments on various topics, such as engineered bacteria, oil-degrading bacteria, and symbiotic relationship between root colonizing bacteria and host plants, have been included here.
Various other innovations in recombinant DNA technology have also been critical constituents of 2021 Microbiology News. So let’s look at research findings from a more comprehensive angle.
Table of Contents
- Top 15 Microbiology News of 2021
- 1. Phages eradicate entirely bacteria that cause dysentery and lessen the virulence capacity of bacteria (USA, Nov 2021)
- 2. Engineered E. coli could produce renewable fuel from carbon dioxide in the form of carbohydrates (USA, Sept 2021)
- Top 14 Biochemistry News, Innovations & Breakthroughs In 2017
- 3. Bacteria associated with plant roots prefer to colonize the roots of their native host plants (Germany, July 2021)
- Top 15 Latest Microbiology & Virology Discoveries in 2018
- 4. Microbial association in kefir makes the impossible possible (U.K, Jan 2021)
- 5. Canadian Arctic marine bacteria are capable of degrading oil and diesel (USA, Aug 2021)
- 6. Effects of climate change on interactions in the microbial network (USA, Feb 2021)
- Field Biologist – How To Become One?
- 7. Researchers identify a novel bacterial trait that may be used to treat and diagnose Lyme disease (USA, Nov 2021)
- 8. Gargle lavage samples can be used in PCR tests to detect SARS-CoV-2 infection just similarly as nasopharyngeal swabs (USA, July 2021)
- 9. Scientists solved the method by which Staphylococci defend themselves from antibiotics (German, Oct 2021)
- 10. Green vegetable sulfosugar encourages the development of crucial gut bacteria (Austria, April 2021)
- 11. Examining the function of microbial predators at hydrothermal vents in the deep sea (USA, July 2021)
- 12. Scientists found on the ocean floor certain specialized bacteria that consume and recycle nucleic acid from dead biomass (Austria, June 2021)
- 13. Scientists found new species of bacteria as ecological bait for sandflies (USA, July 2021)
- 14. Investigators predicted the mechanism as to how archaea consume ethane as their favorite meal (Germany, Sept 2021)
- 15. Food scientists create a road map for deadly Listeria (USA, Aug 2021)
Top 15 Microbiology News of 2021
1. Phages eradicate entirely bacteria that cause dysentery and lessen the virulence capacity of bacteria (USA, Nov 2021)

Phages are bacteria-infecting viruses that are also able to treat infections in humans. However, the use of phages as therapeutics is greatly limited by the ease with which bacteria can develop resistance to phage attacks, much like antibiotics.
- Investigators have now established that Shigella flexneri, a significant cause of dysentery in sub-Saharan Africa and southern Asia, is killed by the naturally occurring phage A1-1, which also favors phage-resistant mutants with decreased virulence.
- The mechanism by which phage enters and kills S. flexneriis by using a specific surface receptor on the bacterium called OmpA, bacteria that survive the phage’s attack will either be devoid of OmpA receptors, or any remaining receptors will have undergone mutations that lessen virulence.
- To find a phage that can naturally bind to the outer membrane proteins of S. flexneriresponsible for the pathogen’s virulent cell-to-cell spread in the human intestine and to evaluate the hypothesis that the evolution of phage resistance will change or eradicate this virulence factor protein.
According to the researchers, S. flexneriwas the target of phage therapy because it was already resistant to common antibiotics. Furthermore, this pathogen primarily affects low-income nations where clean water access and expensive antibiotics are concerns.
Suggested Reading:
Latest Discoveries In Microbiology 2019
![]()
2. Engineered E. coli could produce renewable fuel from carbon dioxide in the form of carbohydrates (USA, Sept 2021)
Escherichia colibacteria have been modified by scientists to capture carbon dioxide and transform it into formic acid using hydrogen gas.
- The production of H2 and CO2 from formic acid is catalyzed by an enzyme in e.coli. The latter is best known in nature as a type of vinegar compound used by ants to fend off predators.
- The researchers forced the bacteria to substitute tungsten for molybdenum, typically an essential enzyme component, by allowing the bacteria to grow in abundance. Following the concept that during the process of evolution absence of oxygen prevailed in the atmosphere three and a half billion years ago, but there were high CO2 and H2, and cellular life had started to evolve 10, 000 meters beneath the ocean’s surface.
- These substances would have needed to be transformed into essential carbohydrates back then. Replacing the cofactors tungsten for molybdenum can change the enzyme activity.
The main objectives of this research would be to capture waste CO2 and convert it to formic acid using electrolysis powered by renewable electricity or renewable hydrogen gas produced from biohydrogen, as in this research. A microbe needs to use formate as its only carbon source. Then one can create chemicals, plastic, or fuel. This illustrates a cyclical bioeconomy in which CO2 is continuously produced, captured, and sold.
Suggested Reading:
Top 14 Biochemistry News, Innovations & Breakthroughs In 2017
![]()
3. Bacteria associated with plant roots prefer to colonize the roots of their native host plants (Germany, July 2021)

According to researchers, bacteria from the plant microbiota are adapted to their host species. They demonstrate how root-associated bacteria can invade a microbiota already developed because they have a competitive advantage when colonizing their native host.
- Roots are a vital point of contact between plants and the soil environment; plants, including crops like rice and wheat, rely on them for essential water and mineral nutrients.
- Land plants’ roots form organized communities known as the root microbiota with various microbes, including bacteria, that are attracted from the surrounding soil. The plant host sustains these microbial communities by giving them nutrients, primarily in the form of organic carbon compounds secreted by the root.
- As a result, these commensal bacteria mediate a few beneficial processes for their plant host, such as supplying pathogen defense, enhancing nutrient mobilization from the soil, and favorably affecting growth.
- The host partially influenced this process by analyzing the gene expression of both plant species when interacting with various synthetic communities. It’s interesting to note that different gene expression profiles for several well-known regulators of plant immunity were present during root colonization by native and non-native SynComs.
- Based on their findings, the authors hypothesized that the creation of species-specific host niches gives native strains a competitive edge when colonizing the roots of their respective host plants.
These findings could significantly impact agriculture because they emphasize the value of bacterial competition and the significance of host preference for successful root colonization. Probiotic inoculants with enhanced capacity to invade and persist in standing microbial communities could aid in overcoming the variation in efficacy of currently used biologicals in agriculture. Furthermore, these inoculants are tailored to specific crop plants.
Suggested Reading:
Top 15 Latest Microbiology & Virology Discoveries in 2018
![]()
4. Microbial association in kefir makes the impossible possible (U.K, Jan 2021)

Scientists know that microorganisms frequently live in communities and depend on one another for survival. Still, their mechanistic understanding of this phenomenon is quite limited.
- Today, many cutting-edge techniques have been combined by researchers to gain a better understanding of microbial communities. This demonstrated that cooperation enables the microbes to accomplish goals they are unable to achieve on their own.
- Members of the group research kefir, one of the oldest fermented foods in the world and an increasingly popular “superfood” with various alleged health advantages, including better digestion, lower blood pressure, and lower blood sugar levels.
- After examining 15 samples of kefir, the researchers were surprised to learn that the predominant species of Lactobacillusbacteria found in kefir grains cannot survive on its own in milk, the other essential component of kefir.
- The species provide for one another’s needs when they cooperate, feeding on each other’s metabolites in the kefir culture. It is fascinating how Lactobacillus kefiranofaciens, the dominant species in the kefir community, assembles all other microbes necessary for survival using kefir grains.
Kefir is a beneficial beverage serving as a model microbial community for studying metabolic interactions. In addition, it is simple to cultivate. While yogurt and kefir are fermented and cultured dairy products rich in “probiotics“, kefir has a much larger microbial community than yogurt, including bacterial and yeast cultures.
Suggested Reading:
Introduction To Fermentation Biology
![]()
5. Canadian Arctic marine bacteria are capable of degrading oil and diesel (USA, Aug 2021)

According to a recently published study, marine bacteria in the arctic waters of Canada can degrade oil and diesel fuel.
- Scientists revealed that by genome sequencing, a specific group of bacterial lineages, including Paraperlucidibaca, Cycloclasticus, and Zhongshania, were capable of hydrocarbon bioremediation.
- As a result, they are considered key players in managing oil spills in the Arctic Ocean. The study also demonstrated that, in these low-temperature conditions, nutrient supplementation can accelerate hydrocarbon biodegradation.
- So, it can be concluded that naturally occurring oil-degrading bacteria serve as nature’s response to oil spills.
Suggested Reading:
What Do Marine Biologists Do?
![]()
6. Effects of climate change on interactions in the microbial network (USA, Feb 2021)

To manage ecosystems and predict the ecological effects of future climate warming, a new study investigates how the complexity and stability of microbial networks are affected by climate change.
- Although soil microbial community diversity, structure, and activities can change due to climate warming, it is still unknown whether and how network complexity and its connections to stability in microbial communities are impacted.
- Therefore, the research team looked at the temporal dynamics of soil microbial communities to determine whether and how climate warming affects the complexity and stability of ecological networks in soil microbial communities.
- Global warming significantly increased the robustness of molecular ecological networks. The strong correlation between network stability and complexity supports the central ecological thesis that complexity breeds stability.
This study makes the case that the stability of the microbial community in the grassland ecosystem and the related ecosystem functions may be less vulnerable in the warmer world. However, climate warming impacts decreased biodiversity and associated ecosystem functioning.
Suggested Reading:
Field Biologist – How To Become One?
![]()
7. Researchers identify a novel bacterial trait that may be used to treat and diagnose Lyme disease (USA, Nov 2021)
According to research, the bacterium that causes Lyme disease, which is common to all bacteria, has a highly unusual modification in its molecular bag of defense.
- The number of cases reported and the geographic distribution of Lyme disease in the United States have dramatically increased over the previous 20 years.
- The Lyme disease-causing bacterium Borrelia burgdorferiis carried by black-legged ticks that live in Virginia and bite humans. Jutras discovered in 2019 that after entering the human body, B. burgdorferireleases peptidoglycan.
- Even though peptidoglycan is present in all bacteria, many do not shed it. Different types of bacteria are the cause of Lyme disease. Its peptidoglycan and its constituent parts are made oddly.
- Chitin, a structural carbohydrate with bound sugar molecules made from modified glucose, is broken down into the sugar that B. burgdorferi incorporates into its peptidoglycan. Ticks must include chitin to survive.
- According to studies, B. burgdorferi’s peptidoglycan stays in the tissues of people with Lyme arthritis long after the bacteria has entered the body. The peptidoglycan is present weeks to months after the initial infection, causing pain and inflammation.
- A protein linked to Borrelia burgdorferi’s peptidoglycan was found in the lab. It acts as a molecular beacon that annoys the patients’ immune systems and amplifies inflammation in Lyme arthritis sufferers.
Given that this is a very novel molecule, this discovery opens up an exciting new field of study for the lab. Researchers believe that these molecules are released by the bacterium, and this finding may pave the way for creating a diagnostic tool that looks for them, much like a biomarker for Lyme disease.
Suggested Reading:
Do Bacteria Have Nucleus?
![]()
8. Gargle lavage samples can be used in PCR tests to detect SARS-CoV-2 infection just similarly as nasopharyngeal swabs (USA, July 2021)

A cohort of 80 individuals was examined for SARS-CoV-2 via nasopharyngeal swab in a newly published study in Microbiology Spectrum, the new open-access journal of the American Society for Microbiology.
- All 26 individuals proved positive using gargle lavage (mouthwash). This is good news because nasopharyngeal swabbing, currently the gold standard for gathering samples for COVID testing, is not widely used because it causes discomfort to the subject.
- The highly sensitive real-time polymerase chain reaction is the method of testing samples used in this study (RT-PCR).
- Our findings demonstrate that the virus could be found in the gargle lavage using the same RT-PCR technique in every instance where a person tested positive by the gold standard nasal swab method.
Based on paired samples from a sizable patient cohort, the finding strongly suggests that the painless self-collection of gargle lavage is a suitable and simple source for accurately detecting SARS-CoV-2.
Suggested Reading:
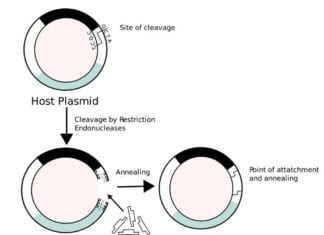
Recombinant Proteins
![]()
9. Scientists solved the method by which Staphylococci defend themselves from antibiotics (German, Oct 2021)
Staphylococcus aureus, a skin bacterium, frequently develops antibiotic resistance. Infections that are challenging to treat may then result from it. Now, scientists have discovered a clever mechanism by which a specific strain of Staphylococcus aureusdefends itself from the critical antibiotic vancomycin.
- Bacteria are lipid-based single-celled organisms that are protected by a thin membrane. The membrane is nearly as delicate as a soap bubble, and the staphylococcal cell’s internal pressure would rupture it.
- Consequently, a cell wall surrounds the membrane, encasing the bacterium like an incredibly durable protective garment. The peptidoglycan, a network of layers of cross-linked carbohydrate chains, makes up this wall.
- Thus, a stable fabric is produced. Staphylococci and other bacteria produce the fundamental components of this fabric inside the cell, which then transport them outside through the membrane.
- They are stopped from becoming part of the wall by the antibiotic vancomycin, which traps them there. Thus, the cells pass away. For instance, autolysins ensure that the cells can separate during cell division by cleaving the cell wall.
- This is why bacteria need them during reproduction. The protective suit’s seams are continually opened by the molecular scissors so that fresh patches of peptidoglycan can be inserted as the cell expands. Autolysins are crucial for this reason.
So thereby, the result shows how Staphylococciadapt to their environment through spontaneous genetic changes and avoid the effects of antibiotics is improved as a result.
Suggested Reading:
Explore 13 Different Shapes of Bacteria
![]()
10. Green vegetable sulfosugar encourages the development of crucial gut bacteria (Austria, April 2021)

A group of researchers has studied how the plant-based sugar sulfoquinovose is broken down by microbes in the gut. Their research revealed that specialized bacteria utilize the sulfosugar, producing hydrogen sulphide. This gas has contradictory effects on human health: while it has an anti-inflammatory effect at low concentrations, higher levels of hydrogen sulfide in the intestine are linked to conditions like cancer.
- Leafy vegetables like spinach might have hundreds of chemical constituents that reach our digestive system. For example, a sulfonic acid derivative of glucose is called sulfoquinovose. It is a chemical building block, primarily in green vegetables such as spinach, lettuce, and algae.
- Unlike glucose, which feeds many microorganisms in the gut, sulfoquinovose stimulates the growth of particular vital organisms in the gut microbiome. These key organisms include the bacterium of Eubacterium rectale, one of the ten most common gut microbes in healthy people.
- Dihydroxypropane sulfonate (DHPS) is one of the products of the metabolic pathway that the E. rectale bacteria use to ferment sulfoquinovose. DHPS is then used as an energy source by other intestinal bacteria.
- After this process, the final product derived is Hydrogen sulphide(H2S). Hydrogen sulfide, among other things, can have an anti-inflammatory effect on the intestinal mucosa in low concentrations.
- On the other hand, increased hydrogen sulfide production by gut microbes is linked to cancer and chronic inflammatory diseases.
Studies revealed that sulfoquinovose can be used to encourage the development of various gut bacteria that are crucial to the health of our gut microbiome. We now understand that these bacteria convert it into the contradictory hydrogen sulphide, concluded researchers. Scientists from Konstanz and Vienna will now conduct additional research to determine whether and how the consumption of sulfosugar derived from plants can positively impact health.
Suggested Reading:
Cellular Organization: Exploring The Cell
![]()
11. Examining the function of microbial predators at hydrothermal vents in the deep sea (USA, July 2021)
A biological hotspot of activity develops in the deep ocean due to the hydrothermal vent fluids from the Gorda Ridge spreading center in the northeast Pacific Ocean.
- In the deep ocean, a unique food web relies on chemical energy from the venting fluids rather than photosynthesis to thrive. Many microbial eukaryotes, or protists, which feed on chemosynthetic bacteria and archaea, have a field day feasting at the Gorda Ridge vents.
- According to the paper, protozoa act as a link between lower trophic levels and primary producers. Their grazing is crucial for carbon transport and recycling in microbial food webs.
- The Gorda Ridge, about 200 kilometers off the coast of southern Oregon, is known for its hydrothermal vents. According to research, protists consume 28 to 62% of the daily stock of bacteria and archaea biomass in these fluids. Additionally, according to their calculations, protistan grazing may be responsible for transferring or consuming up to 22% of the carbon fixed by the chemosynthetic population in the vent fluids.
- The ocean offers us various ecosystem services, including carbon sinks and seafood, that is well-known to many people. However, we don’t have that much information about how those food webs function when we think about microbial ecosystem services, particularly in the deep sea.
This study reveals a vast microbial community and system at work below the euphotic zone, away from the sun’s light.
Suggested Reading:
Top 25 Immune System Fun Facts
![]()
12. Scientists found on the ocean floor certain specialized bacteria that consume and recycle nucleic acid from dead biomass (Austria, June 2021)
Several bacteria that consume nucleic acids have been found in sediment samples from the Atlantic Ocean by an international team of researchers. In fact, one of the team’s newly identified bacteria is a true expert at degrading DNA.
- The team has recently identified one bacterium that is a true expert in destroying DNA. They can utilize various molecules as nutrients, including biomolecules from extinct and decomposing organisms like proteins and lipids.
- This includes so-called extracellular DNA molecules absent from intact cells or no longer present. Kenneth Wasmund, a microbiologist at the Centre for Microbiology and Environmental Systems Science (CMESS) at the University of Vienna and the study’s lead author, claims that DNA is especially nourishing from the perspective of bacteria.
- The sediment bacteria were given purified DNA that was isotopically labeled with heavy carbon atoms (13C). They then tracked the heavy carbon using stable isotope probing, including a particular isotope imaging technique. As a result, they could determine which bacteria broke down the labeled DNA.
- Additionally, to understand the DNA-eating microorganisms’ functional potential and distribution in the world’s oceans, researchers recreated the genetic data found in the cells, or genomes, of the organisms.
- The results of the metagenomic analysis demonstrated that the bacteria had DNA-degrading enzymes that allowed them to break down DNA into smaller pieces for easier uptake and consumption.
- One particular bacterial species stood out because it had a highly developed arsenal of tools for destroying DNA. The research team named them Izemoplasma acidinucleiciin honor of their appetite for DNA, also known as nucleic acid.
So the study predicted the presence of certain novel DNA-eating bacteria with a pool of DNA-degrading enzymes that can be further used for various purposes in recombinant DNA technology.
Suggested Reading:
Are Enzymes Proteins?
![]()
13. Scientists found new species of bacteria as ecological bait for sandflies (USA, July 2021)

While investigating the locations where sandflies raise their young, researchers made an unexpected discovery: a new species of bacteria that is very alluring to sandfly mothers. The research could help develop environmentally friendly baits or traps to control sand fly populations.
- Important parasitic diseases that affect people in tropical and subtropical areas of Asia, Africa, and the Middle East are spread by sandflies. Leishmaniasis is one of them; it typically results in sores and skin ulcers, but it can occasionally harm internal organs.
- Insecticides are difficult to use on sand flies because they hide in sheltered areas like caves, animal burrows, and cracks in rocks or trees.
- Sphingobacterium phlebotomi, a new member of the bacterial family known as Sphingobacteriaceae, was discovered among various bacterial species from the sand fly larval rearing area. A new type of bacteria produces volatile chemicals that sand flies in heat find appealing.
It can be concluded that these new species of bacteria emitting volatile chemicals attracted sand flies that can be used as future remediation against the sand fly populations.
Suggested Reading:
Explore The Cells of The Epidermis
![]()
14. Investigators predicted the mechanism as to how archaea consume ethane as their favorite meal (Germany, Sept 2021)

Microbes that consume ethane live in hot vents in the deep sea. Researchers have now successfully identified a crucial element in the microbial conversion of the gas. The enzyme in charge of fixing ethane was able to have its structure decoded.
- Geothermal heat causes organic matter to deteriorate into oil and natural gases like ethane in deep-sea sediments.
- Archaea, which break down the natural gas, and bacteria, which couple the electrons released in the process to the reduction of sulfate, an abundant compound in the ocean, consume ethane. This group of microorganisms is known as a consortium.
- The discovery of ethane-eating microbes has given the research a new lease on life. The ethane specialists grow much more quickly and double every week compared to microbes that consume methane and take a long time to develop.
- As a result, biomass production takes place over a shorter period, enabling attempts to characterize and purify the essential enzymes that catalyze the oxidation of natural gas.
Although it is a significant advance to discover that the enzyme responsible for the process has unique characteristics that enable it to recognize ethane rather than other alkanes, a complete understanding of the degradation process is still a mechanism to understand.
Suggested Reading:
The Domain Archaea: Finding Life’s Extremists
![]()
15. Food scientists create a road map for deadly Listeria (USA, Aug 2021)

Listeria monocytogenes, one of the most dangerous foodborne pathogens, may soon be simpler to locate in food recalls and other investigations, thanks to a new genomic and geological mapping tool developed by food scientists.
- According to research groups, the objective of this work was to meticulously gather soil samples from all over the country to accurately portray the true large-scale spatial distribution, genomic diversity, and population structure of listeria species in their natural habitat.
- According to Liao, the ecological and evolutionary drivers of bacterial genome flexibility-a significant open question in microbiology-with whole genome sequencing and thorough population genomics analyses.
- By identifying listeria contaminations that might have a natural origin, Liao said that this work will probably help the food industry and can be used as a reference for upcoming population genomics studies.
So, the present study is a blessing as it helps track down the foodborne pathogens responsible for food poisoning.
Suggested Reading:
Explore 9 Stomach Flu Vs Food Poisoning Differences
![]()
This series of microbiology news give us a broader perspective on the recent advancement in this field. Commencing with the discoveries of crucial new species uses of microbial populations as a new field of renewable energy resources, unique concept, and mechanism of gut microbiota, phage therapeutic approaches, and other evolutionary aspects of the microbial world. Through research and innovations, we march forward and anticipate more exciting findings in 2022.
![]()